Slicing a MHD simulation with field lines
[1]:
from pathlib import Path
import imageio
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from matplotlib.colors import LinearSegmentedColormap
from tqdm.auto import tqdm
import astro3d
from astro3d.image_stack import makeslice
from astro3d.image_stack import VeroC_sRGB
from astro3d.image_stack import VeroM_sRGB
from astro3d.image_stack import VeroY_sRGB
from astro3d.image_stack import C_sRGB
from astro3d.image_stack import M_sRGB
from astro3d.image_stack import Y_sRGB
plt.style.use([{'image.cmap':'gray_r'}])
Setup
Read data
[2]:
with np.load(astro3d.get_data('diskdata/rho.npz')) as f:
data = f['arr_0'][()]
with np.load(astro3d.get_output('streamlines.npz'), allow_pickle=True) as f:
streamlines = list(f['pathes'][()])
[3]:
x = np.linspace(0, 1, data.shape[0])
y = np.linspace(0, 1, data.shape[1])
z = np.linspace(0, 1, data.shape[2])
Printer settings
printer specific and layer thickness can be chosen to be different)
[4]:
# these are the values for the J850 Prime
#dpi_x = 600
#dpi_y = 600
#dpi_z = 940 # 0.027 mm layer thickness
# these are the values used in alphacams TEILEFABRIK, where we ordered some of our prints
dpi_x = 600
dpi_y = 300
dpi_z = 940 # 0.027 mm layer thickness = 2.54 / dpi_z
Choose the height of the print, the rest should rescale accordingly
[5]:
height = 5.0 # this should be the total height of the printed cube in cm
We distinguish between Vivid colors and the Rigid Colors. The Vivid colors are transparent and need to be mixed with white or black to make them intransparent.
[6]:
vivid = True
Output folder
We store the images in the path set by output_dir.
[7]:
output_dir = 'slices_mhd' + vivid * '_vivid'
[8]:
path = Path(astro3d.get_output()) / output_dir
if not path.is_dir():
path.mkdir()
else:
files = list(path.glob('slice*.png'))
if len(files)>0:
print('directory exists, deleting old files')
for file in files:
file.unlink()
directory exists, deleting old files
New coordinates
calculate the new grids in x, y, z
[9]:
#n_z = int(height / layer_thickness)
n_z = int(height * dpi_z / 2.54)
n_x = int(n_z * len(x) / len(z) / dpi_z * dpi_x)
n_y = int(n_z * len(y) / len(z) / dpi_z * dpi_y)
n_x += n_x%2 # add 1 to make it even if it isn't
n_y += n_y%2 # add 1 to make it even if it isn't
x2 = np.linspace(x[0], x[-1], n_x)
y2 = np.linspace(y[0], y[-1], n_y)
z2 = np.linspace(z[0], z[-1], n_z)
create an interpolation function for the non-normalized 3D data
[10]:
def f_interp(coords):
return astro3d._fortran.fmodule.interpolate(x, y, z, data, coords)
set the coordinates-tuple which is passed to the interpolation. x and y entries will stay the same and only the z entry will change.
Note that coords will not be passed to the interpolation function in this form, but will be transformed to a shape (N, 3), where the number of points N = len(x2) * len(y2).
[11]:
coords = (x2, y2, z2)
Normalization & color choices
Find the largest magnitude of the data values and define a logarithmic norm
[12]:
dyn_range = 1e-4
vmax = data.max()
norm = LogNorm(dyn_range * vmax, vmax, clip=True)
define the density levels, their width, their colors, and the filling factors of the colors
[13]:
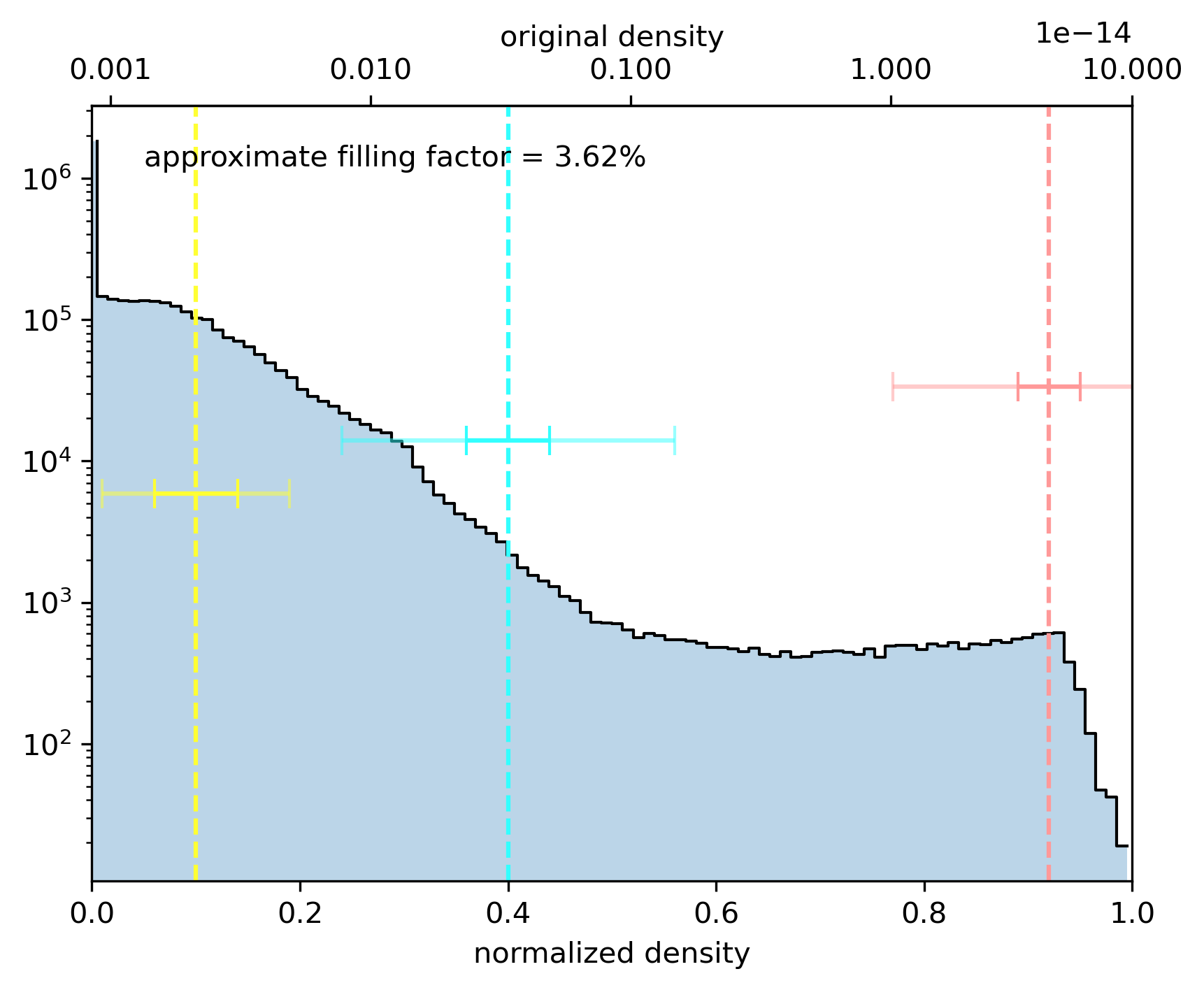
levels = np.array([0.1, 0.4, 0.92])
# sigmas = np.array([0.03, 0.05, 0.03])
# clip = np.array([3.0, 5.0, 5.0])
sigmas = np.array([0.04, 0.04, 0.03])
clip = np.array([2.25, 4.0, 5.0])
# fill = np.array([0.1, 0.2, 1]) # this seemed too full
# fill = np.array([0.03, 0.06, 1.0]) # this seemed too empty
fill = np.array([0.08, 0.14, 0.5]) # also making the disk slightly less solid
We distinguish between Vivid colors and the Rigid Colors. The Vivid colors are transparent and need to be mixed with white or black to make them intransparent.
[14]:
MidGreen = [1, 154, 106]
[15]:
from astro3d.image_stack import rgb_to_cmyk
mg = rgb_to_cmyk(MidGreen)
mg[-1] = 1 - mg[-1]
mg /= mg.sum()
mg = np.round(mg * 100) / 100
[16]:
# we add one extra color for the streamlines
White = np.ones(3)
if vivid:
colors = [[Y_sRGB, White], [C_sRGB, White], [Y_sRGB, M_sRGB, White], [C_sRGB, Y_sRGB, White]]
fmix = [[0.8, 0.2 ], [0.8, 0.2 ], [0.4, 0.4, 0.2 ], [0.58, 0.22, 0.2 ]]
else:
colors = [VeroY_sRGB, VeroC_sRGB, [VeroY_sRGB, VeroM_sRGB], VeroM_sRGB]
fmix = [1, 1, [0.5, 0.5], 1]
[17]:
# show the mixed colors
mix = [(np.array(c, ndmin=2) * np.array(_f, ndmin=2).T).sum(0) for c, _f in zip(colors, fmix)]
plt.imshow([mix]).axes.axis('off');
Show a histogram of the data values
[18]:
astro3d.image_stack.show_histogram(data, norm, colors=colors, levels=levels, sigmas=sigmas, clips=clip, f=fmix, fill=fill)
Example slice
select which index in the new z-grid to process
[19]:
iz = int(np.ceil(n_z / 2))
Process one slice and check the results
[20]:
data_d, img = makeslice(iz, z2, f_interp, coords, norm, path,
levels=levels, sigmas=sigmas, fill=fill,
colors=colors, f=fmix, streamlines=streamlines, bg=0.8)
[21]:
fig, axs = plt.subplots(2, 4, figsize=(16, 8), dpi=300)
axs[0,0].imshow(data[:, :, z.searchsorted(z2[iz])].T,norm=norm)
axs[0,0].text(0.05, 0.95, 'density', va='top', transform=axs[0,0].transAxes)
im = imageio.v2.imread(path / f'slice_{iz:04d}.png')
axs[0,1].imshow(im,norm=norm)
axs[0,1].set_aspect(dpi_x/dpi_y)
axs[0,1].text(0.05, 0.95, 'dithered image', va='top', transform=axs[0,1].transAxes)
axs[0,2].set_visible(False)
axs[0,3].set_visible(False)
for i, ax in enumerate(axs[1,:]):
cmap = LinearSegmentedColormap.from_list('my', [[1,1,1], mix[i]])
ax.imshow(data_d[:, :, i], origin='lower', cmap=cmap)
ax.set_aspect(dpi_x/dpi_y)
ax.text(0.05, 0.95, f'color {i+1}', va='top', transform=ax.transAxes)

Iteration
[22]:
iz = [0] # Here we just want to print the first slice
# iz = np.arange(n_z)
[23]:
for _iz in tqdm(iz):
makeslice(_iz, z2, f_interp, coords, norm, path,
levels=levels, sigmas=sigmas, fill=fill,
colors=colors, f=fmix, streamlines=streamlines, bg=0.5)